Why We Use Sparse Matrices for Recommender Systems
Introduction to SciPy’s Sparse Module
In recommender systems, we typically work with very sparse matrices as the item universe is very large while a single user typically interacts with a very small subset of the item universe. Take YouTube for example — a user typically watches hundreds if not thousands of videos, compared to the millions of videos YouTube has in its corpus, resulting in sparsity of >99%.
This means that when we represent the users (as rows) and items (as columns) in a matrix, the result is an extremely sparse matrix consisting of many zero values (see below).
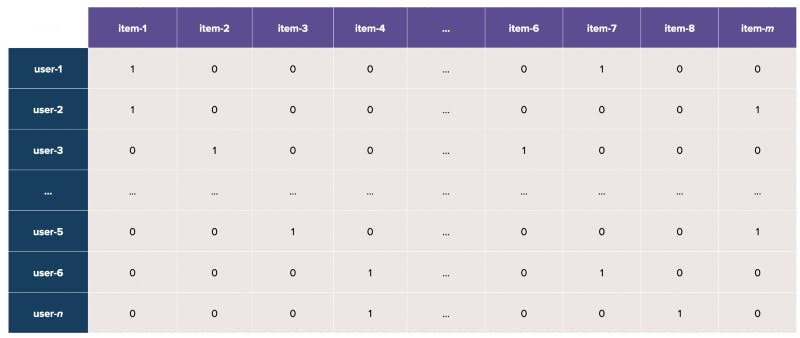
In a real-life scenario, how do we best represent such a sparse user-item interaction matrix?
Why can’t we just use Numpy Arrays or Pandas DataFrames?
To understand this, we have to understand the two major constraints on computing — time and memory. The former is simply what we know as “how much time a program takes to run” whereas the latter is “how much RAM is being used by the program”. The former is quite straightforward but as for the latter, making sure our program doesn’t consume all our memory is important especially when we deal with large datasets, otherwise we would encounter the famous “out-of-memory” error.
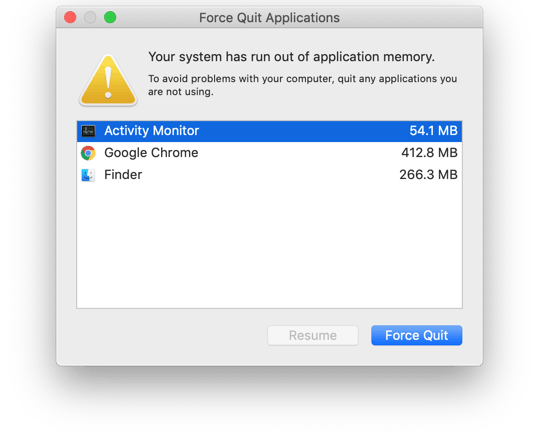
Yes, every program and application on our PC uses some memory (see below image). When we run matrix computations and we want to store those sparse matrices as a Numpy array or Pandas DataFrame, they consume memory as well.
To formalize these two constraints, they are known as time and space complexity (memory).
Space Complexity
When dealing with sparse matrices, storing them as a full matrix (from this point referred to as a dense matrix) is simply inefficient. This is because a full array occupies a block of memory for each entry, so a n x m array requires n x m blocks of memory. From a simple logic standpoint, it simply doesn’t make sense to store so many zeros!
From a mathematical standpoint, if we have a 100,000 x 100,000 matrix, this will require us to have 100,000 x 100,000 x 8 = 80 GB worth of memory to store this matrix (since each double uses 8 bytes)!
Time Complexity
In addition to space complexity, dense matrices exacerbate our runtimes as well. We shall illustrate using an example below.
How then do we represent these matrices?
Introducing… SciPy’s Sparse Module
In Python, sparse data structures are efficiently implemented in the scipy.sparse module, which are mostly based on Numpy arrays. The idea behind the implementation is simple: Instead of storing all values in a dense matrix, let’s just store the non-zero values in some format (e.g. using their row and column indices).
Before we dive into what CSR is, let’s compare the efficiency difference in using numpy arrays versus sparse matrices in both time and space complexity.
import numpy as np
from scipy import sparse
from sys import getsizeof
# **Matrix 1**: *Create a dense matrix (stored as a full matrix).*
A_full = np.random.rand(600, 600)
# **Matrix 2**: *Store A\_full as a sparse matrix (though it is dense).*
A_sparse = sparse.csc_matrix(A_full)
# **Matrix 3**: *Create a sparse matrix (stored as a full matrix).*
B_full = np.diag(np.random.rand(600))
# **Matrix 4**: *Store B_full as a sparse matrix.*
B_sparse = sparse.csc_matrix(B_full)
# Create a square function to return the square of the matrix
def square(A):
return np.power(A, 2)
We then time these different matrices stored in these different forms and also how much memory they use.
%timeit square(A_full)
print(getsizeof(A_full))
>>> 6.91 ms ± 84.5 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
>>> 2880112
%timeit square(A_sparse)
print(getsizeof(A_sparse))
>>> 409 ms ± 11.5 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
>>> 56
%timeit square(B_full)
print(getsizeof(B_full))
>>> 2.18 ms ± 56.5 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
>>> 2880112
%timeit square(B_sparse)
print(getsizeof(B_sparse))
>>> 187 µs ± 5.24 µs per loop (mean ± std. dev. of 7 runs, 10000 loops each)
>>> 56
Clearly, the best performance in terms of both time and space is obtained when we store a sparse matrix with the sparse module.
Compressed Sparse Row (CSR)
Even though there are many types of sparse matrices in SciPy such as Dictionary of Keys (DOK) and List of List (LIL), I will only touch on Compressed Sparse Rows (CSR) because it is the most used and widely known format.
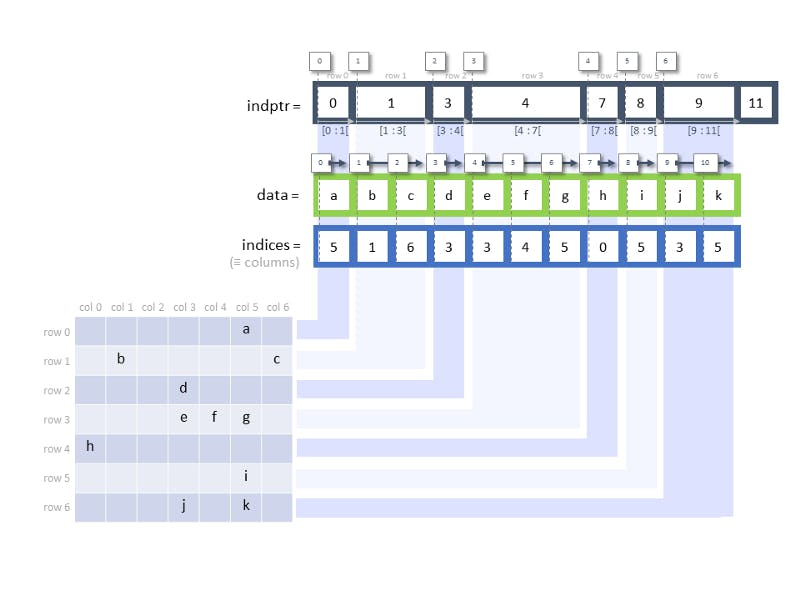
CSR (and also CSC, a.k.a. compressed sparse column) is used for write-once-read-many tasks [1]. To efficiently represent a sparse matrix, CSR uses three numpy arrays to store some relevant information, including:
data: the values of the non-zero values — these are the non-zero values that are stored within the sparse matrixindices: an array of column indices — starting from the first row (from left to right), we identify non-zero positions and return their indices in that row. In the diagram below, the first non-zero value occurs at row 0 column 5, hence 5 appears as the first value in theindicesarray, followed by 1 (row 1, column 1).indptr: stands for index pointer and returns an array of row starts. This definition confuses me and I choose to interpret it this way: it tells us how many of the values are contained in each row. In the below example, we see that the first row contains a single valueaand so we index it with0:1. The second row contains two valuesb, cwhich we then index from1:3and so on. Thelen(indptr) = len(data) + 1 = len(indices) + 1because for each row, we represent it with the start and end index (similar to how we index a list).
What are some ways to construct a csr_matrix?
Create a full matrix and convert it to a sparse matrix
some_dense_matrix = np.random.random(600, 600)
some_sparse_matrix = sparse.csr_matrix(some_dense_matrix)
As seen earlier, this method is not efficient because we have to first obtain this dense matrix which is very memory consuming, before we can convert it into a sparse matrix.
Create an empty sparse matrix
# format: csr_matrix((row_len, col_len))
empty_sparse_matrix = sparse.csr_matrix((600, 600))
Note that we should not create an empty sparse matrix and populate them subsequently because the csr_matrix is designed to write-once-read-many. Writing to the csr_matrix will be inefficient and one should consider other types of sparse matrices like List of lists that is more efficient at manipulating the sparsity structure.
Create a sparse matrix with data
# **method 1**
# format: csr_matrix((data, (row_ind, col_ind)), [shape=(M, N)])
# where a[row_ind[k], col_ind\[k]] = data[k]
data = [3, 9, 5]
rows = [0, 1, 1]
cols = [2, 1, 2]
sparse_matrix = sparse.csr_matrix((data, (rows, cols)),
shape=(len(rows), len(cols))
sparse_matrix.toarray()
>>> array([[0, 0, 3],
[0, 9, 5],
[0, 0, 0]], dtype=int64)
# **method 2**
# format: csr_matrix((data, indices, indptr), [shape=(M, N)])
# column indices for row i: indices[indptr[i]:indptr[i+1]]
# data values: data[indptr[i]:indptr[i+1]]
data = [3, 9, 5]
indices = [2, 1, 2]
indptr = [0, 1, 3, 3]
sparse_matrix = sparse.csr_matrix((data, indices, indptr))
sparse_matrix.toarray()
>>> array([[0, 0, 3],
[0, 9, 5],
[0, 0, 0]], dtype=int64)
Hope this was helpful in getting you kick-started in using sparse matrices!
Support me! — If you like my content, do consider dropping a sponsor :)
